Medical Image Processing & Artificial Intelligence
In Medical Image Processing and Artificial Intelligence researchers at MIBE are working on the development of innovative algorithms for the acquisition, analysis and interpretation of medical image data, in particular using convolutional neural networks, for the future computer-aided diagnosis of diseases.
Computational Imaging
PD Dr. Tobias Lasser
Research group Computational Imaging & Inverse Problems
Publications
"Seeing is understanding" – the goal of Prof. Lasser and his team is to enable them to see more by investigating computational approaches for novel imaging methods. Example applications include:
- visualizing three-dimensional structures, such as a zebrafish brain, from a single planar microscopy image
- as well as identifying microstructures, such as nerve fibers, in macroscopic objects like the human brain.
Using techniques ranging from classical variational methods to modern deep learning approaches, they aim at developing new models and algorithms in computational imaging and inverse problems, in close collaboration with partners from medicine, biology, physics, and mathematics.
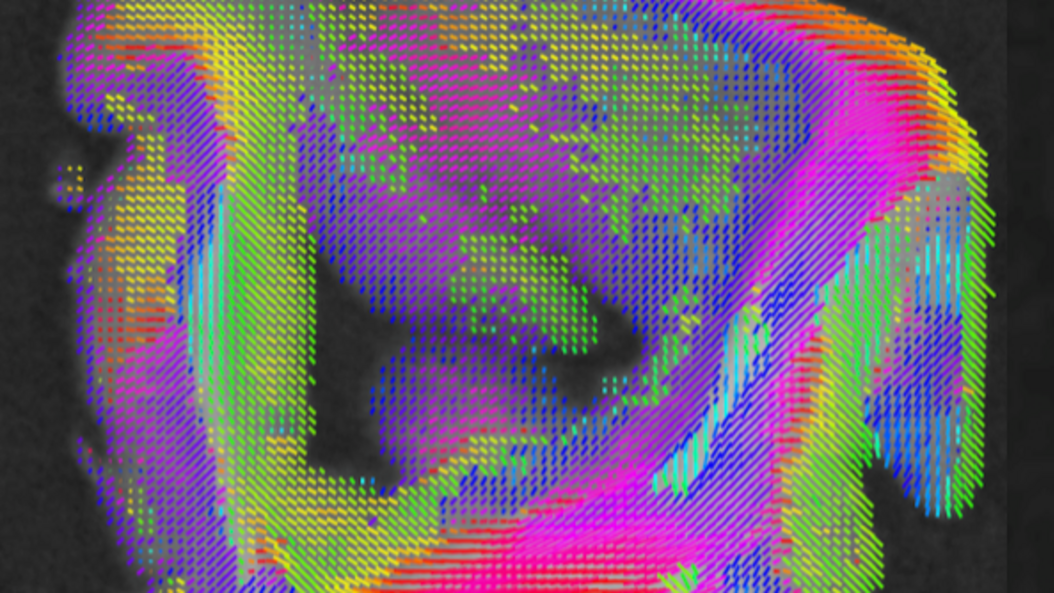
Figure Caption:
Anisotropic X-ray Dark-field Tomography of a human brain sample, showing the microstructure orientations. (Details in Wieczorek et al., Scientific Reports 8, 2018.)
Medical Image Computing
PI Björn Menze
Research Group Biomedical Image Analysis and Machine Learning
Publications
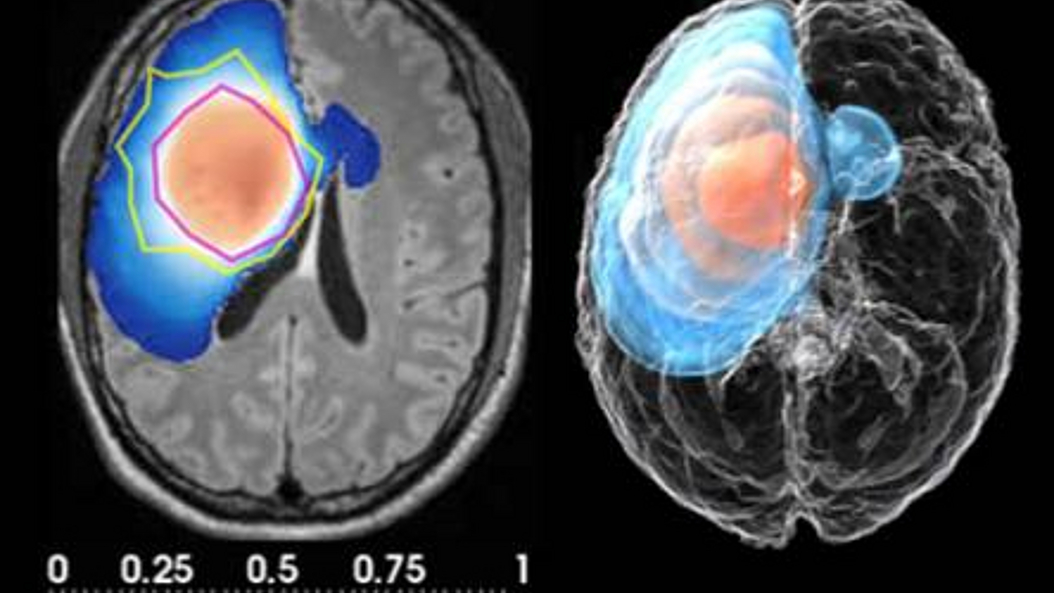
The research of Prof. Menze and his team is in medical image computing, exploring topics at the interface of medical computer vision, image-based modeling, and computational physiology with a strong application focus on oncology. Very broadly, they are interested in developing computational methods that will help in transforming the qualitative visual inspection of medical image data into a functional interpretation of the disease process. In this, they focus (1) on developing principled directions for integrating and abstracting information from complex multi-modal image data by using biophysical-statistical models, and (2) on further developing data-driven machine-learning approaches that provide means for embedding this model-driven analysis in a workflow capable of dealing with large clinical image data sets at the population scale.
In the long term, this work will provide the computational methods necessary to infer function from structure by modeling clinically relevant functional anatomy, and the technical means for developing new (patho-) physiological models at the same pace as novel imaging methods to generate more and more specific insights into anatomy and function.
Computer-aided Medical Procedures
PI Nassir Navab
Chair for Computer Aided Medical Procedures & Augmented Reality
Publications
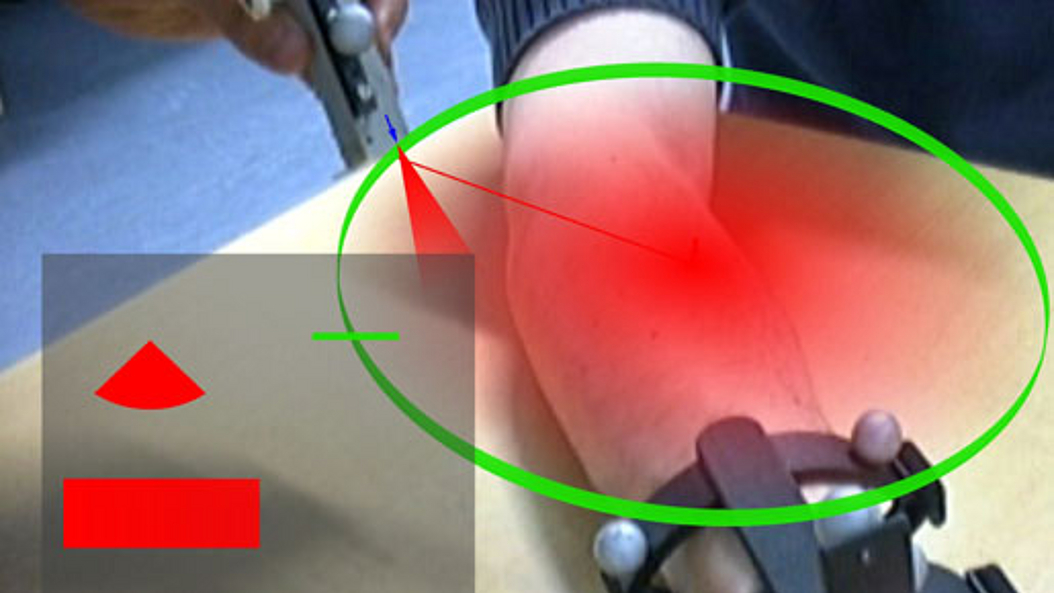
This research project focuses on computer-aided medical procedures and augmented reality. The work involves developing technologies to improve the quality of medical interventions and bridges the gap between medicine and computer science. The research objective is to study and model medical procedures and introduce advanced computer integrated solutions to improve their quality, efficiency, and safety. Prof. Navab and his team aim at improvements in medical technology for diagnosis and therapeutic procedures.
More information:
In the Media: TV feature on medical augmented reality
Cognitive Systems
PI Gordon Cheng
Chair of Cognitive Systems
Publications
The Chair of Cognitive Systems deals with the fundamental understanding and creation of cognitive systems.
Our main research topics are:
- Active Tactile Learning
- Affective Brain-Robot-Interface
- Artificial Robotic Skin
- Cognitive Architectures
- Humanoid Robotic Systems and Locomotion
- Physical-Human Robot Interaction
- Multi-modal Sensor Fusion
- Enhanced Reasoning Methods
- Self-aware Robots
- Social Robotics
More Information:
YouTube Channel
HPC Modeling of Bio-Materials
PI Wolfgang A. Wall
Chair of Computational Mechanics
Publications
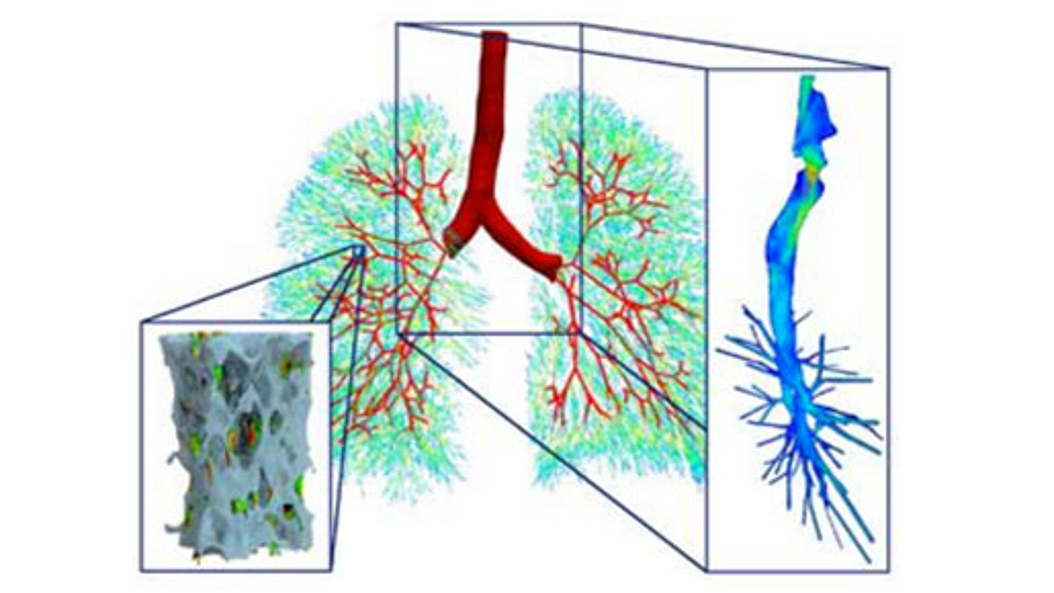
The knowledge about the basic biophysical properties of materials and interaction on the microscopic and mesoscopic scale can be used to computationally model healthy properties and certain diseases in tissues organs. One such example is mechanical ventilation for patients suffering from lung diseases. Here, computer simulations can predict so-called ventilation-induced lung injuries, by modeling the airflow from the windpipe to the smallest airways and the resulting local straining of the tissue. Information obtained from these very complex simulations can help physicians predict the effect of different interventions and develop improved patient-specific treatments in the future.
Healthcare & Rehabilitation Robotics
Prof. Dr. Cristina Piazza
Assistant Professorship of Healthcare and Rehabilitation Robotics
Publications
The loss of an upper limb severely affects a person’s life and autonomy. In some situations, prostheses can offer valuable assistance to recover some of the lost functionalities. Cristina Piazza, Professor of Healthcare and Rehabilitation Robotics, and her team develop and improve bionic prostheses such as robotic hands and wearable devices. These systems are inspired by neuroscientific principles and exploit emerging technologies, i.e. the use of soft robotics, to overcome current functional and technical limitations and improve the performance of these devices. In addition, the team explores methods and advanced algorithms based on myoelectric control, to translate user intentions in commands for the prosthesis. Moreover, the researchers develop virtual reality environments that can be used to evaluate the perception of the prosthesis and develop further technologies.
Bioinformatics & Computer-aided Biology
Prof. Dr. Burkhard Rost
Professorship of Bioinformatics
Publications
Professor Rost conducts research on bioinformatics and computer-aided biology, with a focus on predicting the functions and structures of proteins and genes. His particular interest is predicting protein interactions and the effects of changing individual amino acids, with the goal of fostering a better understanding of how proteins, genes and cells work. He also focuses on enabling earlier diagnosis and more effective treatment of illnesses. The specific niche of his research group links artificial intelligence and machine learning to evolution.
Mathematical Modelling of Biological Systems
Prof. Dr. Fabian J. Theis
Professorship of Mathematical Modelling of Biological Systems
Publications
Fabian Theis conducts research in the field of computational biology. The main focus of his work is the application of machine learning methods to biological questions, in particular as a means of modeling cell heterogeneities on the basis of single cell analyses and also of integrating “omics” data into systems medicine approaches.
Computational Neurosciences
Prof. Dr. Julijana Gjorgjieva
Computational Neurosciences
Publikationen
Professor Gjorgjieva (b. 1983) conducts research in the fields of computational and theoretical neuroscience. She is interested in how brain circuits become tuned to maintain a balance between constant change as we learn new things, and robustness to produce reliable behavior. In particular, she concentrates on two aspects of neural circuit organization, looking at how it emerges from the interaction of neuronal and synaptic properties during development, and from optimality and energy conservation principles that operate over the longer timescales of evolution.
Computer Vision & Artificial Intelligence
Prof. Dr. Daniel Cremers
Chair of Computer Vision & Artificial Intelligence
Publications
Computer vision, machine learning & deep networks, mathematical image analysis (segmentation, motion estimation, multiview reconstruction, visual SLAM), shape analysis, autonomous systems & self-driving cars, variational methods and partial differential equations, convex and combinatorial optimization & statistical inference.
More Information:
Watch his recent TEDx talk on 3D Computer Vision. And public lectures on Machine Intelligence for Self-Driving Cars, on Past and Future of Artificial Intelligence, and on Recent Advances in 3D Shape Analysis.